각종 샘플(세포, 조직, 환경샘플 등)로부터 고품질
DNA나 RNA를 가장 쉽고 빠르게 뽑을 수 있습니다.
이 외에도, 각종 Epigenetics 관련 제품들
(DNA Methylation kit 등)과 Microbiomics
(샘플 채집부터 분석까지의 전 단계의 제품)
연관 제품들이 준비되어 있습니다.
제품정보
Kyongshin PRODUCT

제품소개


| Equipment Needed (user provided) | Thermal cycler with heated lid, magnetic stand for 0.2 mL PCR tubes, microcentrifuge for 0.2 mL PCR tubes and 1.5 mL microcentrifuge tubes, and a benchtop vortex mixer. A complimentary magnet stand is available at online checkout for direct U.S. customers of R3000 and R3003. |
|---|---|
| Input Quality | RNA should be free of DNA contamination and enzymatic inhibitors, with A260/A280 and A260/A230 ≥ 1.8. RNA with lower purity ratios (A260/A280 and A260/A230) should be treated with DNase I and purified with the RNA Clean & Concentrator™ (Cat. No. R1013) prior to processing. RNA should be suspended in water, TE, or a low-salt buffer.
|
| Library Storage | Libraries eluted in DNA Elution Buffer (provided) may be stored at ≤ 4°C overnight or ≤ -20°C for long-term storage. |
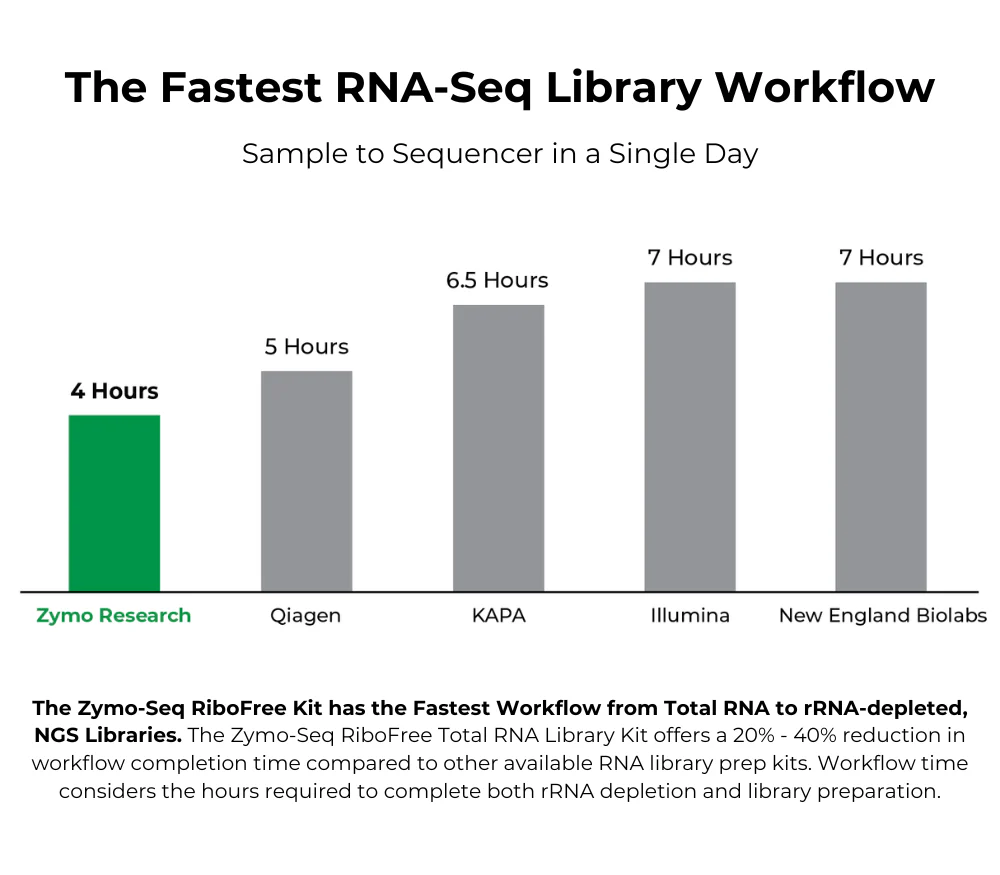
| Processing Time | As little as 4 hours |
| RNA Input | 10 – 250 ng of total RNA.
|
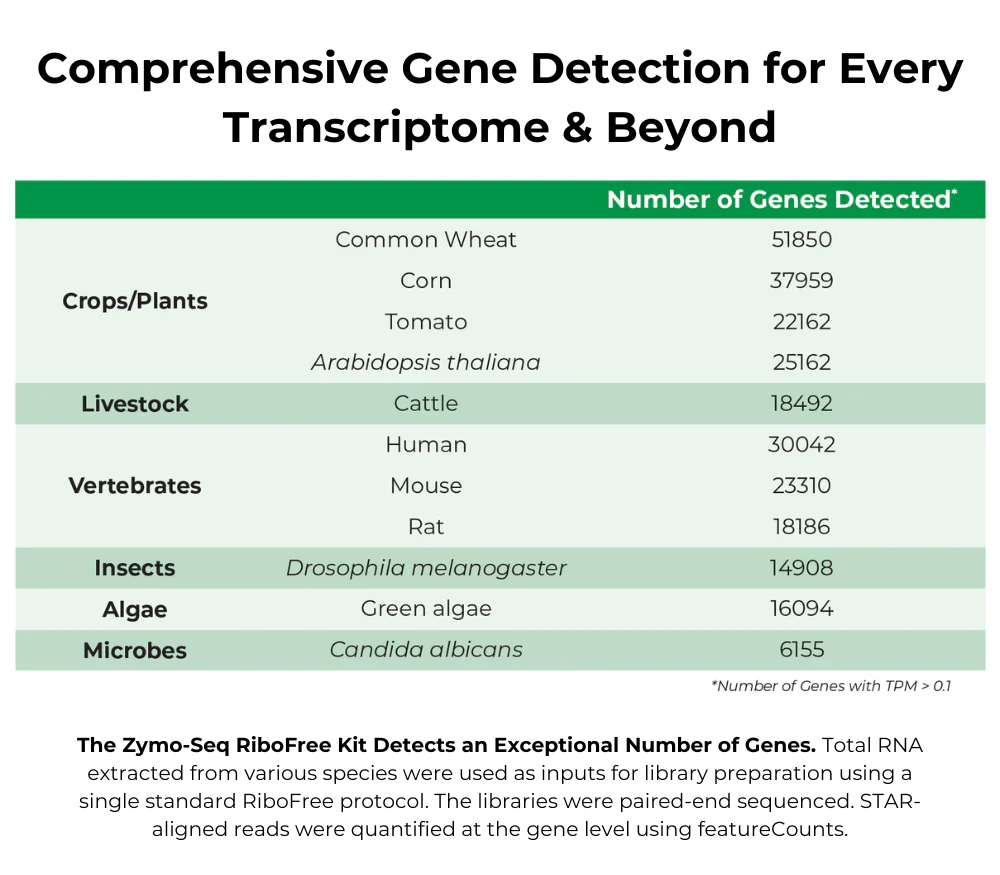
| Sample Input Material | RNA from any species |
| Sequencing Platform Compatibility | Libraries are compatible with all Illumina® sequencing platforms except HiSeq® X. Libraries are compatible with the Element AVITI™ system. While technically possible, Illumina® originally limits the applications on HiSeq® X exclusively for whole-genome libraries. Please confirm with the sequencing service provider for acceptability and additional details if expecting to sequence Zymo-Seq RiboFree Total RNA libraries on HiSeq® X series sequencers. |
주문정보
| CAT.No | 품명 | 규격 | 비고 |
|---|---|---|---|
| R3000 | Zymo-Seq RiboFree Total RNA Library Kit | 12 preps | |
| R3003 | Zymo-Seq RiboFree Total RNA Library Kit | 96 preps |